Ancient Genome Browser
A High Coverage Denisovan Genome
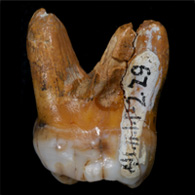
The genome sequence of a Denisovan individual was generated from a small fragment of a finger bone discovered in Denisova Cave in southern Siberia in 2008.
Approximately 30-fold coverage of the genome was generated using the Illumina GAIIx sequencing platform.
This data is freely available without passwords.
Focussing on the "mappable genome" - the 1.86GB of unique sequence with clear human-chimpanzee orthology - we see 99.97% of bases covered at least 3x and 99.51% covered at least 10x.
Contamination with modern human DNA, estimated from both mitochondrial and nuclear genomes, is less than 1%.
Downloads
The Denisova Genome Consortium have released for public use the raw sequence data, alignments and genotype calls for the high-coverage genome sequence of a Denisovan individual, as well as the modern human sequences presented in the publication.
When using this genome data please cite the following publication:
Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, Sudmant PH, Alkan C, Fu Q, Do R, Rohland N, Tandon A, Siebauer M, Green RE, Bryc K, Briggs AW, Stenzel U, Dabney J, Shendure J, Kitzman J, Hammer MF, Shunkov MV, Derevianko AP, Patterson N, Andres AM, Eichler EE, Slatkin M, Reich D, Kelso J, Paabo S: A High-Coverage Genome Sequence from an Archaic Denisovan Individual.Science. Aug 31 2012.
Provided here are the alignments using BWA to the human (hg19/GRCh37) and chimpanzee (panTro2) genomes.
Download data
The sequence, alignment and genotype data is made available through:
- local site: http://cdna.eva.mpg.de/denisova/
- Amazon Web Services: The alignments and raw sequence data are available as a public data set via Amazon Web Services (AWS) at http://aws.amazon.com/datasets/2357.
- UCSC Genome Browser


Last updated: May 19, 2015