Adam Powell

Senior Researcher
Max Planck Institute for Evolutionary Anthropology
Department of Human Behavior, Ecology and Culture
Deutscher Platz 6
04103 Leipzig
E-mail: adam_powell@[>>> Please remove the text! <<<]eva.mpg.de
Office: Level 1, Room B1.11
Research interests
Curriculum Vitae
Software
Publications
Research interests
I work on the interactions between human genetic and cultural evolutionary systems, integrating methods and data from population genetics, archaeogenetics, archaeology, anthropology and historical linguistics to understand human population history. I develop integrated simulation and statistical inference methods, aiming to build a unified gene-culture coevolutionary framework for human demographic inference. I am a member of the Theory in Cultural Evolution Lab based in HBEC, and lead the ERC-funded research project Waves, focusing on the population history of the South Pacific.
Curriculum Vitae
EDUCATION & EMPLOYMENT
| 2019 to present | Senior Scientist / ERC Research Group Leader Department of Human Behaviour, Ecology and Max Planck Institute for Evolutionary Anthropology (MPI-EVA), Leipzig, Germany |
| 2015 to 2019 | Research Group Leader: Gene-culture Coevolution Department of Linguistic and Cultural Evolution & Department of Archaeogenetics Max Planck Institute for the Science of Human History (MPI-SHH), Jena, Germany |
| 2013 to 2015 | Postdoctoral Research Associate Palaeogenetics Group, Institute of Anthropology, Johannes Gutenberg University, Mainz, Germany |
| 2010 to 2012 | Postdoctoral Research Associate UCL Genetics Institute, University College London (UCL), UK |
| 2006 to 2010 | PhD in Population genetics “Demography and the evolution of genetic and cultural variation” Department of Genetics, Evolution and Environment (GEE), University College London (UCL), London, UK |
| 1999 to 2003 | BSc in Mathematics (Honours) University of Exeter, Exeter, UK |
GRANTS & AWARDS
| 2018 | Waves of history in the South Pacific: A gene-culture coevolutionary approach. European Research Council (ERC) Starting Grant (758967), PI, €1,500,000 |
| 2018 | INTERACT: Human interactions during the Mesolithic-Neolithic transition in Western Europe: The dual perspectives of cultural and biological exchanges. ANR-DFG, PIs Marie-France Deguilloux & Dr Wolfgang Haak, co-investigator, €671,562 |
| 2018 | Waves of Change: Human migration and adaptation in prehistoric Indonesia. Marsden Fund, New Zealand (18-UOO-135), PI Dr Rebecca Kinaston, associate investigator, NZD 300,000 |
| 2018 | Em busca de pegadas invisíveis: para um modelo multidisciplinar de difusão das línguas Khoe-Kwadi e do pastoralismo na África Austral. Fundação para a Ciência e a Tecnologia, Portugal (29273), PI Jorge Rocha, co-investigator, €237,464 |
| 2010 | 1st prize award, Postgraduate Symposium, Department of Genetics, Evolution and Environment, UCL |
| 2006 | 4-year PhD studentship, Arts and Humanities Research Council (AHRC) Centre for the Evolution of Cultural Diversity, UCL |
Software
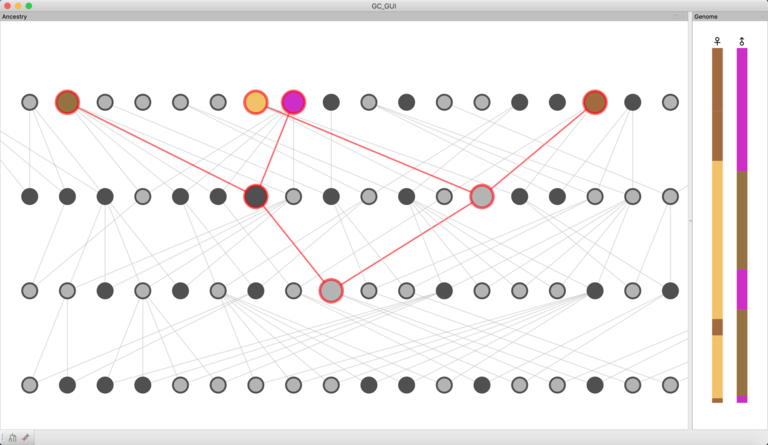
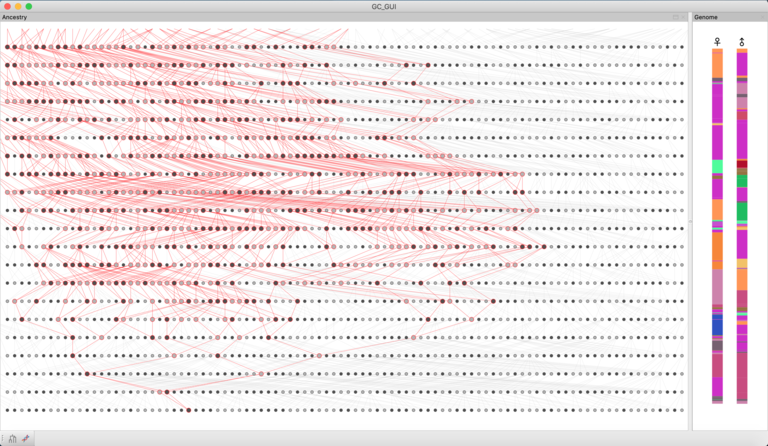
DeMIGOD (Designing Models for Inference in Gene-culture Or Demography) is a gene-culture coevolution simulation platform, combining methods from GIS, ecology, archaeology, population genetics, phylogenetics and cultural evolution into a single unified framework for modeling population prehistory. Its flexible GIS UI allows for the creation and seamless integration of archaeological, topographic and /or palaeoenvironmental data to condition demographic models, which can include population competition, spatiotemporally restricted interactions and cultural boundary formation. Given a simulated history, DeMIGOD will generate expected genetic data (on the genomic-scale), cultural data or both. These simulated datasets can be used alongside observed data to test between various models of population history, as well as to test the power of existing inference methods. Written in C++, OpenGL and wxWidgets. Full release in 2023.
KinGLy (Kinship and Genetic Legacy) is an exploratory gene-culture simulation tool for quantifying the effects of cultural structures and processes – such as kinship systems, cousin-marriage practices or polygamy – on observable genetic or genomic measures, such as runs of homozygosity (RoH). As part of a collaboration with BirthRites IMPRG Leader Dr Heidi Colleran, KinGLy is now being developed into a generative empirical tool for testing the limits to inferring past social systems from archaeological and genomic data. Built in C++ and wxWidgets.
Publications
Miszkiewicz, J. J., Buckley, H. R., Feldman, M., Kiko, L., Carlhoff, S., Nägele, K., Bertolini, E., Guimarães, N. R. D., Walker, M. M., Powell, A., Posth, C., & Kinaston , R. L. (2022). Female bone physiology resilience in a past Polynesian Outlier community. Scientific Reports, 12: 18857. |
|
Oliveira, S., Nägele, K., Carlhoff, S., Pugach, I., Koesbardiati, T., Hübner, A., Meyer, M., Oktaviana, A. A., Takenaka, M., Katagiri, C., Murti, D. B., Putri, R. S., Mahirta, Petchey, F., Higham, T., Higham, C., O’Connor, S., Hawkins, S., Kinaston, R., Bellwood, P., Ono, R., Powell, A., Krause, J., Posth, C., & Stoneking, M. (2022). Ancient genomes from the last three millennia support multiple human dispersals into Wallacea. Nature Ecology & Evolution, 6, 1024-1034. |
|
Fogarty, L., Ammar, M., Holding, T. M., Powell, A., & Kandler, A. (2022). Ten simple rules for principled simulation modelling. PLoS Computational Biology, 18(3): e1009917. |
Carlhoff, S., Duli, A., Nägele, K., Nur, M., Skov, L., Sumantri, I., Oktaviana, A. A., Hakim, B., Burhan, B., Syahdar, F. A., McGahan, D. P., Bulbeck, D., Perston, Y. L., Newman, K., Saiful, A. M., Ririmasse, M., Chia, S., Hasanuddin, Pulubuhu, D. A. T., Suryatman, Supriadi, Jeong, C., Peter, B. M., Prüfer, K., Powell, A., Krause, J., Posth, C., & Brumm, A. (2021). Genome of a middle Holocene hunter-gatherer from Wallacea. Nature, 596, 543-547. |
Posth, C., Nägele, K., Colleran, H., Valentin, F., Bedford, S., Gray, R. D., Krause, J., & Powell, A. (2019). Response to “Ancient DNA and its contribution to understanding the human history of the Pacific Islands” (Bedford et al. 2018). Archaeology in Oceania, 54, 57-61. |
Kandler, A., & Powell, A. (2018). Generative inference for cultural evolution. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences, 373(1743): 20170056. |
|
Posth, C., Nägele, K., Colleran, H., Valentin, F., Bedford, S., Kami, K. W., Shing, R., Buckley, H., Kinaston, R., Walworth, M., Clark, G. R., Reepmeyer, C., Flexner, J., Maric, T., Moser, J., Gresky, J., Kiko, L., Robson, K. J., Auckland, K., Oppenheimer, S. J., Hill, A. V. S., Mentzer, A. J., Zech, J., Petchey, F., Roberts, P., Jeong, C., Gray, R. D., Krause, J., & Powell, A. (2018). Language continuity despite population replacement in Remote Oceania. Nature Ecology & Evolution, 2(4), 731-740. |
Unterländer, M., Palstra, F., Lazaridis, I., Pilipenko, A., Hofmanová, Z., Groß, M., Sell, C., Blöcher, J., Kirsanow, K., Rohland, N., Rieger, B., Kaiser, E., Schier, W., Pozdniakov, D., Khokhlov, A., Georges, M., Wilde, S., Powell, A., Heyer, E., Currat, M., Reich, D., Samashev, Z., Parzinger, H., & Burger, V. I. M. &. J. (2017). Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe. Nature Communications, 8: 14615. |
Henrich, J., Boyd, R., Derex, M., Kline, M. A., Mesoudi, A., Muthukrishna, M., Powell, A., Shennan, S. J., & Thomas, M. G. (2016). Understanding cumulative cultural evolution. Proceedings of the National Academy of Sciences of the United States of America, 113(44), E6724-E6725. |
|
Posth, C., Renaud, G., Mittnik, A., Drucker, D. G., Rougier, H., Cupillard, C., Valentin, F., Thevenet, C., Furtwängler, A., Wißing, C., Francken, M., Malina, M., Bolus, M., Lari, M., Gigli, E., Capecchi, G., Crevecoeur, I., Beauval, C., Flas, D., Germonpré, M., van der Plicht, J., Cottiaux, R., Gély, B., Ronchitelli, A., Wehrberger, K., Grigorescu, D., Svoboda, J., Semal, P., Caramelli, D., Bocherens, H., Harvati, K., Conard, N. J., Haak, W., Powell, A., & Krause, J. (2016). Pleistocene mitochondrial genomes suggest a single major dispersal of non-Africans and a late glacial population turnover in Europe. Current Biology, 26(6), 827-833. |
Kandler, A., & Powell, A. (2015). Inferring learning strategies from cultural frequency data. In A. Mesoudi, & K. Aoki ( |
|
Scheu, A., Powell, A., Bollongino, R., Vigne, J.-D., Tresset, A., Çakırlar, C., Benecke, N., & Burger, J. (2015). The genetic prehistory of domesticated cattle from their origin to the spread across Europe. BMC Genetics, 16: 54. |
Kamberov, Y. G., Wang, S., Tan, J., Gerbault, P., Wark, A., Tan, L., Yang, Y., Li, S., Tang, K., Chen, H., Powell, A., Itan, Y., Fuller, D., Lohmueller, J., Mao, J., Schachar, A., Paymer, M., Hostetter, E., Byrne, E., Burnett, M., McMahon, A. P., Thomas, M. G., Lieberman, D. E., Jin, L., Tabin, C. J., Morgan, B. A., & Sabeti, P. C. (2013). Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell, 152(4), 691-702. |
|
Bollongino, R., Nehlich, O., Richards, M. P., Orschiedt, J., Thomas, M. G., Sell, C., Fajkošová, Z., Powell, A., & Burger, J. (2013). 2000 years of parallel societies in Stone Age Central Europe. Science, 342(6157), 479-481. |
|
Gavin, M., Botero, C. A., Bowern, C., Colwell, R. K., Dunn, M., Dunn, R. R., Gray, R. D., Kirby, K. R., McCarter, J., Powell, A., Rangel, T. F., Steppe, J. R., Trautwein, M., Verdolin, J. L., & Yanega, G. (2013). Towards a mechanistic understanding of linguistic diversity. Bioscience, 63, 524-535. |
Gerbault, P., Powell, A., & Thomas, M. G. (2012). Evaluating demographic models for goat domestication using mtDNA sequences. Anthropozoologica, 47(2), 64-76. |
|
Bollongino, R., Burger, J., Powell, A., Mashkour, M., Vigne, J.-D., & Thomas, M. G. (2012). Modern Taurine cattle descended from small number of Near-Eastern founders. Molecular Biology and Evolution, 29(9), 2101-2104. |
|
Brace, S., Barnes, I., Powell, A., PEarson, R., Woolaver, L. G., Thomas, M. G., & Turvey, S. T. (2012). Population history of the Hispaniolan hutia Plagiodontia aedium (Rodentia: Capromyidae): Testing the model of ancient differentiation on a geotectonically complex Caribbean island. Molecular Ecology, 21(9), 2239-2253. |
Gerbault, P., Liebert, A., Itan, Y., Powell, A., Currat, M., Burger, J., Swallow, D. M., & Thomas, M. G. (2011). Evolution of lactase persistence: An example of human niche construction. Philosophical Transactions of the Royal Society B: Biological Sciences, 366(1566), 863-877. |
Veeramah, K. R., Connell, B. A., Pour, N. A., Powell, A., Plaster, C. A., Zeitlyn, D., Mendell, N. R., Weale, M. E., Bradman, N., & Thomas, M. G. (2010). Little genetic differentiation as assessed by uniparental markers in the presence of substantial language variation in peoples of the Cross River region of Nigeria. BMC Evolutionary Biology, 10: 92. |
Itan, Y., Powell, A., Beaumont, M. A., Burger, J., & Thomas, M. G. (2009). The origins of lactase persistence in Europe. PLoS Computational Biology, 5(8): e1000491. |
|
Powell, A., Shennan, S., & Thomas, M. G. (2009). Late pleistocene demography and the appearance of modern human behavior. Science, 324(5932), 1298-1301. |
|
Powell, A., Thomas, M. G., & Shennan, S. J. (2009). Demography and variation in the accumulation of culturally inherited skills. In M. J. O'Brian, & S. J. Shennan ( |
 Open Access
Open Access