Population Genetics
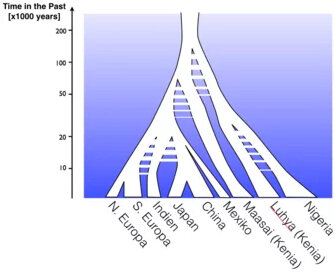
Our group analyses genetic data from ancient and modern populations to advance our understanding of human history.
Examples include the inference of historical population size changes within the last 200,000 years, exploring world-wide diversification of populations after the migration out of Africa within the last 50,000 years, and studying particularly recent migration events, such as the Anglo-Saxon migration period into England 1500 years ago. Particular focus is put on the development of mathematical models and new methods to push the limits of genetic inference towards the more recent past.
Group members

Stephan Schiffels
(Group Leader)

Selina Carlhoff
(Postdoc)

Thiseas Christos Lamnidis
(Postdoc)

Angela Mötsch
(Postdoc)

Luka Papac
(Postdoc)

Lei Huang
(Doctoral Student)

Laura Lacher
(Doctoral Student)
Research Areas
Computational Methods for Genomic Analyses
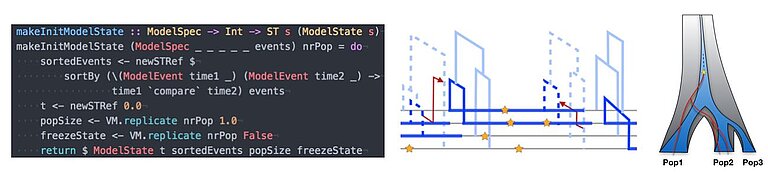
An important part of our work consists of developing new methods to analyse genomic data in order to statistically infer details about the underlying evolutionary dynamics. At the heart of this methodological work is the growing amount of data available in the field of population genomics. When in 2010 there were only a handful of high quality human genomes completely sequenced, nowadays it is thousands and growing. The information that this data carries about our past demography, population diversification, and adaptation processes is vast, in principle. However, extracting this information out of genomes requires novel methods that are both based on sound foundations of population genetic theory and efficient enough to cope with the Big Data that the field of Genetics has become. Furthermore, a relatively novel requirement of such methods is the integration of ancient DNA, which is becoming increasingly available. Important examples of methods that I have developed in recent years and which are further developed in my group include MSMC (published in Schiffels and Durbin, 2014) and rarecoal (published in Schiffels et al. 2016 and Flegontov et al. 2017). We are also very active in developing bioinformatic tools and pipelines to help with processing sequencing data from ancient DNA. Examples for such tools include the sequenceTools package, and various processing tools in the rarecoal-tools package, and the Poseidon-Framework.
Human History through genetics
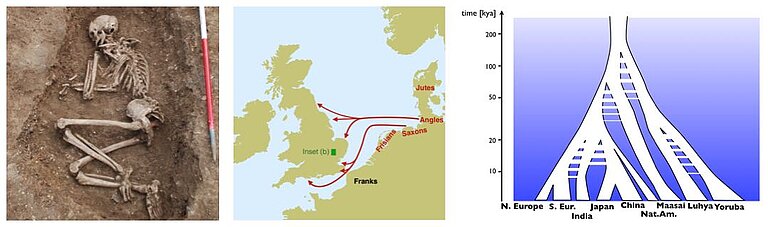
One important goal of genomic analysis is to reconstruct human history by mean of genetics. Up until a few years ago, this was an endeavour that was mostly based on modern genomic data that is became increasingly available. As a major game changer, we are now able to not only indirectly look into the past through our population genetic models based on modern data, but directly through analysing ancient DNA. This relatively new field of "Genetic history" has lead to new insights into human history from Genetics in recent years, including on migration movements between regions and continents, the separation of people into spatially separated subpopulations, as well as changes in population size. Concrete examples of our research covers continental peopling events, for example into America (see Raghavan et al. 2015 and Flegontov et al. 2017), Australia (see Malaspinas et al. 2016), the early Anglo-Saxon migrations into Britain (see Schiffels et al. 2016 and two blog posts here and here), or insights into population changes in Egypt within the last 2,000 years. Our work has also been picked up by the press, for example by the BBC or the Washington Post.
Population Genetic Theory
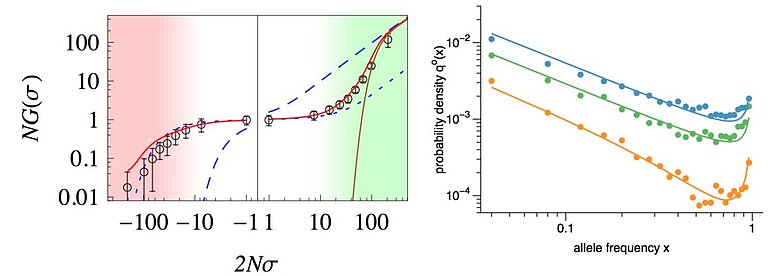
I received my PhD from the Institute for Theoretical Physics in Cologne, and my dissertation was in population genetic theory. Since then I have worked on a number of studies that explore evolutionary dynamics of adaptation, in particular in low recombining organisms or genomic regions. Examples for such studies include the effect of clonal interference on fixation probabilities of selected mutations (see Schiffels et al. 2011), how quantitative trait equilibria affect the the segregations of mutations at a genomic level (see Nourmohammad et al. 2012), and analyses of adaptive evolution under strong linkage in natural populations of Drosophila melanogaster (Schiffels et al. 2017).
Affiliated Researchers
- Joscha Gretzinger (Now Postdoc in the Genetic History Group)
- Elina Salmela
- Suzanne Freilich
Alumni
- Ke Wang
- Ezgi Altınışık
- Martina Čížková
- Tina Saupe
- Margherita Vanni
